Abstract
Aims: The European Collaborative Project on Inflammation and Vascular Wall Remodeling in Atherosclerosis – Intravascular Ultrasound (ATHEROREMO-IVUS) study aims to investigate the relations of genetic profile and novel circulating biomarkers with coronary plaque phenotype and vulnerability as determined by intravascular ultrasound (IVUS).
Methods and results: ATHEROREMO-IVUS is a prospective, observational cohort study of 846 patients with stable angina pectoris or acute coronary syndrome (ACS) who are referred for coronary angiography. Prior to the catheterisation procedure, blood samples are drawn for biomarker measurements and genetic analyses. During the catheterisation procedure, IVUS is performed in a non-culprit coronary artery. The primary endpoint is the presence of vulnerable plaque as determined by IVUS virtual histology. Secondary endpoints include the incidence of major adverse cardiac events during long-term follow-up.
Conclusions: Results from ATHEROREMO-IVUS are expected to improve our knowledge of the role of genetic profile and circulating biomarkers in relation to the development of atherosclerosis and vulnerable plaques. Assessment and early validation of the prognostic value of novel biomarkers and intracoronary imaging techniques will be performed. (ClinicalTrials.gov number: NCT01789411).
Introduction
Coronary artery disease is projected to become the largest single cause of disease burden worldwide1. The traditional view that atherosclerosis is simply a lipid storage disease has evolved, taking into consideration the growing body of evidence that genetic profile, inflammation and blood coagulation play a pivotal role in all stages of atherosclerotic disease, from endothelial dysfunction to late-stage plaque rupture2-5. Genetic markers and circulating biomarkers of inflammation, lipids and coagulation may potentially improve risk stratification in patients with atherosclerotic cardiovascular disease, since they provide information on the biological processes in individuals6,7. Furthermore, these markers may also have a role in the development of new therapeutic targets. Genome-wide scanning of single nucleotide polymorphisms (SNPs) and plasma lipidomics are two potential methods to identify novel genetic and lipid-related markers of coronary artery disease. In vivo intracoronary imaging may further improve coronary risk stratification. Intravascular ultrasound (IVUS) backscattering analysis allows for in vivo differentiation of various plaque phenotypes and may therefore be well suited for detection of plaques that are at high risk of rupture8,9.
The European Collaborative Project on Inflammation and Vascular Wall Remodeling in Atherosclerosis –Intravascular Ultrasound (ATHEROREMO-IVUS) is designed as an exploratory (non-pivotal) clinical study to investigate the associations between genetic profile, circulating biomarkers and coronary atherosclerosis phenotype and vulnerability as determined by IVUS virtual histology. Additionally, novel intracoronary imaging techniques, including near-infrared spectroscopy (NIRS), will be explored to identify lipid core plaques in the coronary arterial wall10. Finally, the prognostic implications of (the combination of) established and novel biomarkers and plaque phenotypes will be studied.
Methods
TARGET POPULATION
The ATHEROREMO-IVUS target population consists of patients with stable angina pectoris or acute coronary syndrome (ACS) who are referred for coronary angiography. The inclusion and exclusion criteria are presented in Table 1. Stable angina pectoris was defined as having at least two of the following three criteria: 1) substernal chest discomfort of characteristic quality and duration; 2) provoked by exertion or emotional stress; 3) relieved by rest and/or glyceryl trinitrate11. ACS includes ST-segment elevation myocardial infarction (STEMI), non-ST-segment elevation myocardial infarction (NSTEMI) and unstable angina pectoris. STEMI was defined as ischaemic symptoms, persistent (>20 min) ST-segment elevation in two contiguous electrocardiogram (ECG) leads and a rise in cardiac enzymes12. Patients with acute chest pain and a typical rise and fall in cardiac enzymes but without persistent ST-segment elevation were classified as NSTEMI. Unstable angina was defined as acute or worsened chest pain without persistent ST-segment elevation and without elevated cardiac enzymes13.

STUDY SAMPLE
The ATHEROREMO-IVUS study cohort mainly consists of patients who were enrolled between 2008 and 2011 in the Erasmus MC, Rotterdam, The Netherlands, which is an academic tertiary referral hospital serving a population of approximately 1.9 million. This cohort was enriched with eligible patients who participated in the Integrated Biomarker and Imaging Study-2 (IBIS-2) trial of darapladib versus placebo (inclusion period 2005-2006)14.
The ATHEROREMO-IVUS study was approved by the medical ethics committee of the Erasmus MC. The study was performed in accordance with the criteria described in the Declaration of Helsinki. Written informed consent was obtained from all enrolled patients.
BLOOD SAMPLING
Blood samples were collected to enable genome-wide scans, lipid mass spectrometry and the analysis of circulating biomarkers. Blood samples were drawn from the arterial sheath prior to the coronary angiography or percutaneous coronary intervention (PCI) procedure. Blood samples were transported to the local clinical chemistry laboratory for further processing (i.e., centrifugation followed by serum, citrate- and EDTA-plasma aspiration and buffy coat separation from the EDTA tube) and storage at a temperature of –80°C within two hours.
GENOME-WIDE SCANS
Genome-wide scans are performed to identify a set of genetic variants that correlate with the extent and phenotype of coronary atherosclerosis. The Affymetrix GeneChip® Human Mapping 6.0 Array (Affymetrix, Santa Clara, CA, USA) is used for the genome-wide scans of 906,600 SNPs. Quality control was performed, including correction for population structure, removal of related samples or samples with mismatched gender.
LIPID EXTRACTION AND MASS SPECTROMETRY
An aliquot of plasma or serum is subjected to lipid extraction. Known amounts of internal standards are added to the samples before extraction and the final lipid extracts are dried under nitrogen. The extracts are reconstituted as described elsewhere15. Sphingolipids are analysed on a 4000 QTRAP® mass spectrometer (Applied Biosystems/MDS Analytical Technologies, Life Technologies, Paisley, UK) equipped with an ultra-high pressure liquid chromatography (UHPLC) system, CTC PAL autosampler (Leap Technologies, Carboro, NC, USA) and Rheos Allegro UHPLC (Flux Instruments AG, Reinach, Switzerland) using multiple reaction monitoring16. Shotgun lipidomics is performed by multiple precursor ion and neutral loss scanning on a QTRAP® 5500 mass spectrometer (Applied Biosystems/MDS Analytical Technologies, Life Technologies) equipped with a robotic nanoflow ion source NanoMate HD (Advion, Ithaca, NY, USA)17. Mass spectrometry data files are processed using MultiQuant™ 1.1.0.26 or Lipid Profiler™ (Applied Biosystems/MDS Analytical Technologies, Life Technologies)18. Identified lipids are quantified by normalising against their respective internal standard and tissue wet weight for aorta and volume for plasma. Quality control (QC) samples are utilised to monitor the overall quality of the lipid extraction and mass spectrometry analyses19. The QC samples are mainly used to remove technical outliers and lipid species that are detected below the lipid class-based lower limit of quantification (LLOQ).
INTRAVASCULAR IMAGING
Following the standard coronary angiography, eligibility for intracoronary imaging was assessed. IVUS data were acquired in a non-culprit coronary vessel. The order of preference for selection of the non-culprit vessel was: 1) left anterior descending (LAD) artery; 2) right coronary artery (RCA); 3) left circumflex (LCX) artery. All IVUS data were acquired with the Volcano s5/s5i Imaging System (Volcano Corp., San Diego, CA, USA) using a Volcano Eagle Eye Gold IVUS catheter (20 MHz). An automatic pullback system was used with a standard pullback speed of 0.5 mm per second.
A number of selected patients in the Erasmus MC also participated in the ATHEROREMO-NIRS substudy (Online Appendix). In these patients, NIRS was performed in the same segment of the non-culprit vessel.
IVUS VIRTUAL HISTOLOGY
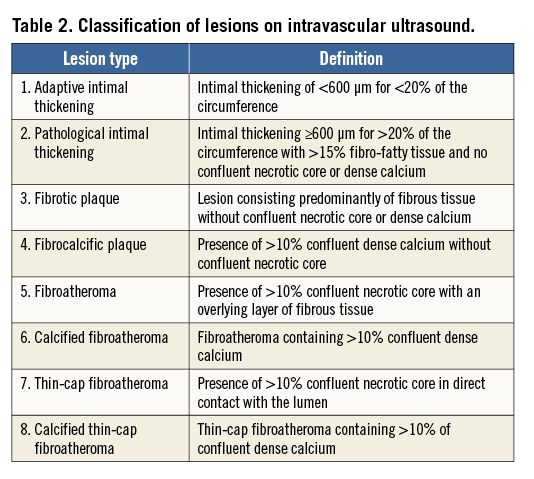
The IVUS greyscale and IVUS radiofrequency backscatter analyses, also known as IVUS virtual histology, were performed using pcVH 2.1 and qVH (Volcano Corp.) software. The baseline IVUS images were analysed offline in an independent core laboratory (Cardialysis BV, Rotterdam, The Netherlands). The core laboratory personnel were blinded for baseline patient characteristics as well as for biomarker, genetic and clinical outcomes data. The external elastic membrane and luminal borders were contoured for each frame (median interslice distance, 0.40 mm). Extent and phenotype of the atherosclerotic plaque were assessed. Plaque burden was defined as the plaque and media cross-sectional area divided by the external elastic membrane cross-sectional area. A coronary lesion was defined as a segment with a plaque burden of more than 40% in at least three consecutive frames (Figure 1). Using IVUS radiofrequency analyses, the composition of the atherosclerotic plaque was characterised into four different tissue types: fibrous, fibro-fatty, dense calcium and necrotic core8. In consensus sessions with three investigators who were blinded to the patient characteristics and outcomes, the lesions were further classified into different lesion types (Table 2)9. A thin-cap fibroatheroma (TCFA) lesion was defined as a lesion with presence of >10% confluent necrotic core in direct contact with the lumen. Remodelling of a lesion was assessed by means of the remodelling index, expressed as the external elastic membrane cross-sectional area at the site of minimal luminal area divided by the reference external elastic membrane cross-sectional area. The reference site was selected <10 mm proximal to the lesion, with no major side branches between the site of the minimal luminal area and the reference.
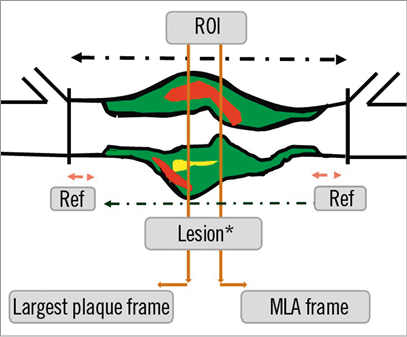
Figure 1. Methodology for detection of lesion and reference segments. MLA: minimal luminal area; Ref: reference segments; ROI: region of interest
FOLLOW-UP
Clinical follow-up started at inclusion and will last for at least one year. Post-discharge survival status will be obtained from municipal civil registries. Post-discharge rehospitalisations will be prospectively assessed during follow-up. Questionnaires focusing on the occurrence of major adverse cardiac events (MACE) will be sent to all living patients. Treating physicians and institutions will be contacted for additional information whenever necessary. If possible and if clinically relevant, culprit and non-culprit-lesion-related events will be distinguished. The occurrence of MACE will be adjudicated by an independent clinical events committee on the basis of original source data and without knowledge of other patient, biomarker, genetic or intracoronary imaging characteristics.
STUDY ENDPOINTS
The primary objective of ATHEROREMO-IVUS is to correlate genetic markers and circulating (lipid) biomarkers with coronary plaque phenotype as determined by IVUS virtual histology. Therefore, the primary endpoint is defined as the presence of TCFA lesions on the imaged non-culprit coronary segment.
The secondary objective is to assess the prognostic value of established biomarkers, novel genetic and lipid biomarkers and plaque phenotypes as determined by IVUS virtual histology. Therefore, the secondary endpoint is defined as the one-year incidence of MACE, which includes all-cause mortality, ACS or unplanned coronary revascularisation. All-cause mortality was defined as death due to any cause20. ACS was defined as the clinical diagnosis of STEMI, NSTEMI or unstable angina pectoris in accordance with the guidelines of the European Society of Cardiology21. Unplanned coronary revascularisation was defined as unplanned repeat PCI or coronary artery bypass grafting (CABG) due to progressive angina or ACS.
SAMPLE SIZE
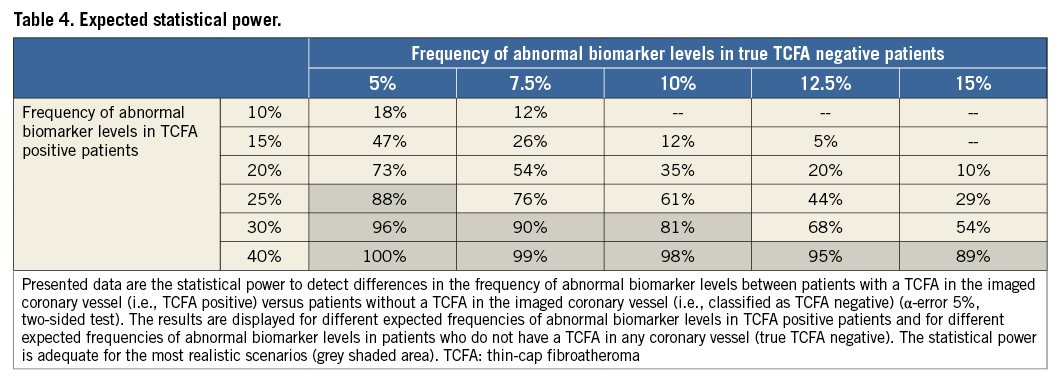
ATHEROREMO-IVUS is designed to explore multiple relations. Its sample size is fixed at 800 patients, which is sufficient to reveal relations between “abnormal” biomarkers (of any kind) and the presence of TCFA lesions with reasonable statistical certainty. We acknowledge that confirmatory studies might be required to establish more firmly relations that we will discover.
Based on prior studies, we expected that 30% of the patients would have a TCFA lesion in at least one coronary artery (i.e., TCFA positive), while 70% of the patients would not have any TCFA lesion (i.e., true TCFA negative) (Figure 2)9. Since TCFA lesions are more or less randomly distributed across the coronary system, we expected that 10% of the patients would have a TCFA lesion in the imaged coronary artery (i.e., classified as TCFA positive), while 90% of the patients would not have a TCFA lesion in the imaged coronary artery (i.e., classified as TCFA negative). In patients who are classified as TCFA negative, 77.8% are expected to be true TCFA negative.
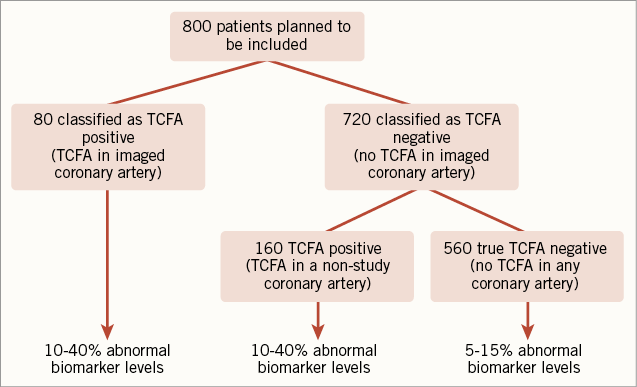
Figure 2. Expected prevalence of thin-cap fibroatheroma lesions. TCFA: thin-cap fibroatheroma
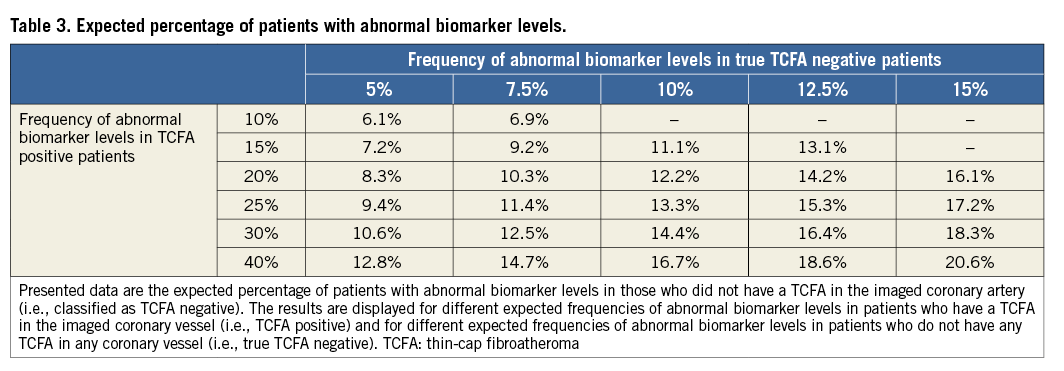
A biomarker level in the upper quintile of its sample distribution is considered as “abnormal”. We expect to observe abnormal biomarker levels in 5-15% of the true TCFA negative patients and in 10-40% of the TCFA positive patients. Hence, the proportion of abnormal biomarker levels in the patients who are classified as TCFA negative is expected to range from 6.1-20.6% (Table 3). Table 4 presents the statistical power to detect differences in the frequency of abnormal biomarker levels between patients who are classified as TCFA negative versus those who are classified as TCFA positive (α-error 5%, two-sided test). The power is adequate (≥80%) for the most realistic scenarios.
Power calculations were not performed for the secondary endpoint. Based on the results of other studies and previous registries in our hospital, we expect that MACE will occur in 5-10% of the patients within the first year of follow-up22-25.
Statistical analyses
Conventional linear regression will be applied to relate SNPs, sphingolipids and other biomarkers with IVUS virtual histology measures, corrected for segment length. Mixed linear models will be used for per-lesion analyses. The relation between biomarkers (in a broad sense) and clinical endpoints will be studied by Cox proportional hazard models.
The p-values that appear in the analyses of genetic variants and sphingolipids will be corrected for multiple testing with appropriate methods to adjust for inflation of the type I error (e.g., Bonferroni or simulation). Significant SNPs and lipid fractions need (and will be proposed for) validation in different datasets.
Actual inclusion
A total of 846 patients with complete data for genetic and lipidomics analyses are included in ATHEROREMO-IVUS: 581 patients were enrolled in the Erasmus MC and 265 participated in IBIS-2 (Figure 3)14. Blood samples are available for 1,098 patients. NIRS was performed in 203 patients.
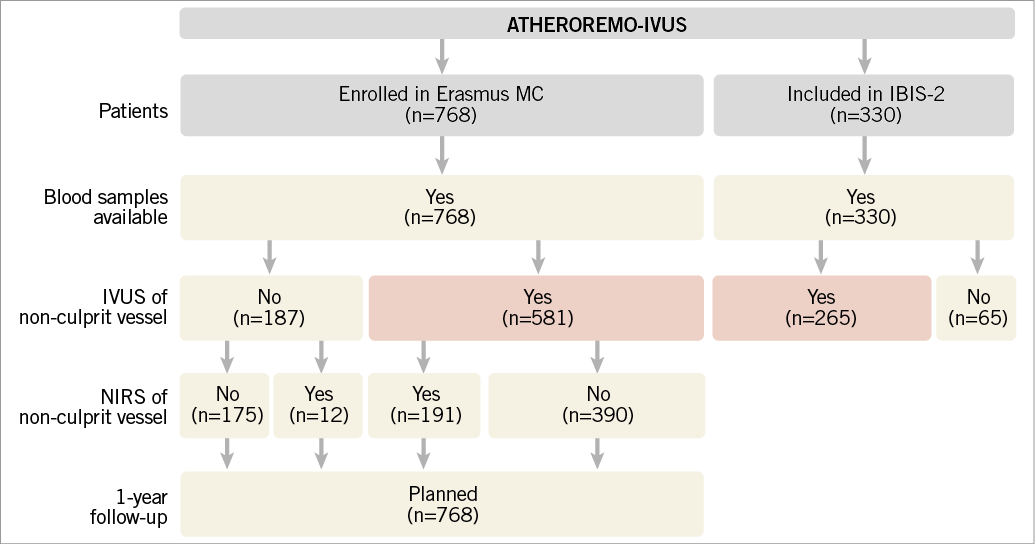
Figure 3. Actual patient inclusion. Finally, blood samples were stored for 1,098 patients, IVUS of a non-culprit vessel was performed in 846 patients (red shaded) and NIRS was performed in 203 patients. IBIS-2: Integrated Biomarker and Imaging Study-2; IVUS: intravascular ultrasound; NIRS: near-infrared spectroscopy
Discussion
The ATHEROREMO-IVUS study was primarily designed to assess correlations of genetic profile and novel circulating biomarkers with the extent, phenotype and vulnerability of coronary atherosclerotic plaques as determined in vivo by IVUS. Furthermore, we would like to assess the potential prognostic value of novel biomarkers, IVUS and NIRS compositional features of atherosclerotic plaques in major cardiac events at long-term follow-up.
Acute coronary syndromes are mostly caused by rupture of TCFA lesions that contain a lipid-rich necrotic core covered by a thin fibrous cap2,26-29. IVUS virtual histology may be suitable for the detection of such vulnerable plaques. The PROSPECT study has shown that TCFA lesions as identified in vivo by IVUS virtual histology were associated with an increased risk for recurrent cardiovascular events in ACS patients. However, the events in PROSPECT were driven mainly by rehospitalisations for unstable or progressive angina, while less is known about the prognostic value of IVUS virtual histology for acute cardiac events as a consequence of spontaneous plaque rupture (i.e., recurrent ACS or death). The prognostic value of IVUS virtual histology in patients with stable angina also remains unclear. Furthermore, the prognostic value of NIRS for the occurrence of MACE has not yet been investigated. The results of the ATHEROREMO-IVUS study will provide data on these questions.
Coronary artery disease has a strong genetic component. Epidemiological studies suggest that up to 50% of its susceptibility is heritable30. Genome-wide scans may measure hundreds of thousands of SNPs that can be tested for an association with a coronary atherosclerosis. Although this method has been shown to be successful in identifying genetic associations with complex traits31, genotyping research programmes for atherosclerosis have been of limited importance so far. One of the bottlenecks was the phenotypic complexity of atherosclerotic vascular diseases. The ATHEROREMO-IVUS study is therefore regarded as a unique opportunity to link genotypes with extensive intracoronary imaging data that reach far beyond the limited knowledge of luminal patency (or stenosis) from conventional coronary angiography.
Several biomarkers of inflammation, coagulation, myocardial necrosis and neurohumoral activation (e.g., C-reactive protein, high-sensitive troponin-T and natriuretic peptides) have more or less been established27. Our aim is to explore novel lipid biomarkers in the first instance, while validation of more established biomarkers will be done in a later stage of the study.
The design of the ATHEROREMO-IVUS study has several strengths. To the best of our knowledge, this is the first (large-scale) study to combine several novel intracoronary imaging techniques with extensive genetic analyses, biomarker exploration and validation and adverse clinical outcome during follow-up. Secondly, in this study we examine a single non-culprit vessel. Ex vivo as well as in vivo studies using IVUS in patients with myocardial infarction have demonstrated the presence of TCFAs in other than the culprit lesion or even culprit artery5. Our approach will allow us to test the hypothesis that the phenotype of a non-culprit artery segment (indicating the patient’s atherosclerotic disease burden) can be linked to biomarker, genetic and outcome data. If the imaging characteristics of the non-culprit artery appear to be related to the incidence of MACE, then this can be seen as a confirmation that the non-culprit artery reflects the atherosclerotic disease burden of the larger coronary vasculature.
Limitations
Some limitations of this study have to be acknowledged. Firstly, the genetic profile and biomarkers will be correlated with the phenotype of the imaged non-culprit coronary artery only. Although we expect that the presence of TCFA lesions is randomly distributed through the coronary system, we may miss the patient’s dominant phenotypic characteristic if this phenotype is only expressed in a coronary segment that has not been imaged (e.g., culprit lesion). Secondly, the ATHEROREMO-IVUS study was designed to explore and discover new genetic and circulating biomarkers. Newly discovered SNPs and lipid biomarkers remain to be validated in another patient cohort.
Conclusions
The results from the ATHEROREMO-IVUS study will improve our understanding of the role of genetic profile and circulating biomarkers in the development of atherosclerosis and vulnerable plaques. Genome-wide scans and lipidomics may identify novel biomarkers in coronary artery disease. Furthermore, the prognostic value of novel circulating biomarkers as well as in vivo detection of vulnerable plaques by IVUS virtual histology and NIRS will be assessed. These findings may further contribute to the improvement of risk assessment in patients with coronary artery disease, which may be important for the optimal choice of treatment in the individual patient.
Guest Editor
This paper was Guest Edited by Franz-Jozef Neumann, MD, Universitäts-Herzzentrum Freiburg-Bad Krozingen, Klinik für Kardiologie und Angiologie II, Bad Krozingen, Germany.
Acknowledgements
The ATHEROREMO-IVUS study is embedded in The European Collaborative Project on Inflammation and Vascular Wall Remodeling in Atherosclerosis (ATHEROREMO) and is funded by the Seventh Framework Programme (FP7), theme FP7-HEALTH-2007-2.4.2-1.
The authors thank Professor Wim van der Giessen, who unfortunately passed away shortly before preparing this manuscript, for his valuable contribution to the study.
Funding
The IBIS-2 study was funded by a research grant from GlaxoSmithKline.
Conflict of interest statement
The authors have no conflicts of interest to declare. The Guest Editor has no conflict of interest to declare.
Appendix. Details of the ATHEROREMO-NIRS substudy
BACKGROUND
Near-infrared spectroscopy (NIRS) is a novel intracoronary imaging technique that may detect lipid core plaques in the coronary arterial wall32,33. Therefore, it may be suitable for in vivo detection of vulnerable plaques. Currently, no data are available on the long-term prognostic value of NIRS in patients with coronary artery disease (CAD). Furthermore, associations between NIRS measurements, genetic markers and circulating biomarkers have not yet been investigated. However, these associations may further elucidate the pathophysiology of coronary lipid core plaques.
OBJECTIVES
The primary objective of the ATHEROREMO-NIRS substudy is to correlate genetic markers and circulating biomarkers with coronary plaque phenotype as determined by NIRS. The secondary objective is to assess the prognostic value of NIRS in patients who underwent coronary angiography for stable angina pectoris or acute coronary syndrome.
METHODS
Prior to coronary angiography, a total of 203 patients provided written informed consent for enrolment in the ATHEROREMO-NIRS substudy, in which NIRS imaging was performed in a single, non-stenotic segment of a non-culprit coronary artery. The order of preference for selection of the non-culprit vessel was: 1) left anterior descending artery; 2) right coronary artery; 3) left circumflex artery. The FDA-approved NIRS system, as used in this study, consists of a 3.2 Fr rapid exchange catheter, a pullback and rotation device and a console (InfraReDx, Burlington, MA, USA). Image acquisition is performed by a motorised catheter pullback at a speed of 0.5 mm/sec and 240 rpm in a proximal segment of a non-culprit artery, starting distal to a side branch. The system performs one thousand chemical measurements per 12.5 mm, in which each measurement interrogates one to two mm2 of vessel wall from a depth of approximately 1 mm in the direction from the luminal surface towards the adventitia1,2. Tissue scattering and absorption of light in the NIR region result in a wavelength-dependent return of light to optical detectors that produces a spectrum. Areas of the artery with spectral characteristics of lipid core are displayed as an image map (chemogram) of the studied vessel.
Yellow regions in the chemogram represent high probability for the presence of lipid core-containing coronary plaques (LCP), while red regions represent those with low probability. The x-axis of the chemogram indicates the pullback position in millimetres, while the y-axis indicates the circumferential position of the measurement from zero to 360 degrees. The lipid core burden index (LCBI) score is computed on the basis of the chemogram by multiplying the fraction of valid yellow pixels by 1,000. Hence, LCBI is a summary measure of the amount of LCP along the entire imaged section of the coronary artery on a 0-to-1,000 scale. NIRS images are analysed offline by an independent core laboratory (Cardialysis BV, Rotterdam, The Netherlands). Core laboratory personnel are blinded to all other baseline patient, biomarker, genetic and outcome data.
Major adverse cardiovascular events, defined as the composite of all-cause mortality, non-fatal ACS, stroke and unplanned coronary revascularisation during one-year follow-up, were adjudicated by a clinical events committee (CEC) on the basis of original source data. Members of the CEC were blinded to other patient data, NIRS imaging characteristics and genetic or biomarker information.

