- myocardial infarction
- STEMI
- cytokines
- gene expression
Abstract
Aims: Transcriptome patterns associated with acute myocardial infarction at the site of coronary occlusion are largely unknown. The aim of this study was to decipher the angiogenic, atherosclerotic, and inflammatory mRNA profiles in whole blood samples collected at the site of coronary occlusion in patients with ST-elevation myocardial infarction (STEMI).
Methods and results: In five consecutive patients with STEMI, blood was sampled at the site of occlusion (local) and in the systemic circulation (peripheral) during primary percutaneous coronary intervention. RNA was extracted from whole blood samples. Among 221 genes involved in angiogenesis, inflammation and atherosclerosis, 24 were shown to be differentially modulated locally, by analysis with custom-designed DNA array technology. Validation in 28 distinct STEMI patients using real-time quantitative PCR identified seven out of these 24 genes to be consistently and significantly upregulated in local versus peripheral blood (p<0.05). Three genes were chemokine family members (CCL2, CCL18 and CXCL12), three genes belonged to the cell-cell and cell-extracellular matrix family (FN1, CDH5 and SPP1), and one gene was representative of the lipoprotein family (APOE).
Conclusions: We identified a set of whole blood transcripts induced at the site of coronary occlusion in the acute phase of myocardial infarction. Resolved genes indicate a predominant role for chemokines, cell-extracellular matrix, and lipoprotein alterations in the pathophysiology of acute myocardial infarction and the initial response to myocardial injury.
Abbreviations
C4A complement component 4A (Rodgers blood group)
CCL18 chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated)
CCL2 chemokine (C-C motif) ligand 2
CCL21 chemokine (C-C motif) ligand 21
CCL24 chemokine (C-C motif) ligand 24
CCR6 chemokine (C-C motif) receptor 6
CCR7 chemokine (C-C motif) receptor 7
CCR9 chemokine (C-C motif) receptor 9
CXCR1 chemokine (C-X3-C motif) receptor 1
IL17C interleukin 17C
IL1B interleukin 1, beta
LTBR4 leukotriene B4 receptor
SPP1 secreted phosphoprotein 1
TOLLIP toll interacting protein
ANGPTL3 angiopoietin-like 3
BAI1 brain-specific angiogenesis inhibitor 1
CDH5 cadherin 5, type 2 (vascular endothelium)
COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen)
FGFR3 fibroblast growth factor receptor 3
APOE apolipoprotein E
EGR1 early growth response 1
FN1 fibronectin 1
PDGFRB platelet-derived growth factor receptor, beta polypeptide
PPARG peroxisome proliferator-activated receptor gamma
Introduction
Atherosclerosis is a chronic disease characterised by inflammatory infiltrates, lipid accumulation, cell death and fibrosis1. Experimental evidence demonstrates the pivotal role of a bimodal inflammatory and neoangiogenic response mediated by white blood cells in all stages of atherosclerosis, from initiation through progression of disease, as well as in plaque rupture and thrombosis2,3. Such pathological processes within the atherosclerotic artery lead to increased serological levels of inflammatory cytokines and related acute-phase reactants, such as C-reactive protein, interleukin-6, tumour necrosis factor-alpha1, as well as anti-inflammatory cytokines such as interleukin-104. This inflammatory pattern is characteristic of the “biomarker panel” found in patients with acute coronary syndrome5. Indeed, preliminary human studies have reported elevated levels of inflammatory biomarkers such as interleukin-6, serum amyloid A, myeloid related protein 8/14 (MRP8/14) and Toll-like receptors-4 (TLR-4)6-8 at the site of coronary occlusion supporting their involvement in the pathophysiology of plaque rupture in acute myocardial infarction (AMI)7.
The identification of biomarkers reflective of plaque destabilisation remains the unmet clinical need. Whole blood gene expression analysis in the acute phase of myocardial expression may help to reveal not only critical factors participating in plaque rupture but identify novel biological targets. In fact, there is no information on the transcriptome pattern of inflammatory or angiogenic response during the acute phase of human myocardial infarction at the site of the coronary occlusion in human AMI. Accordingly, the aim of this study was to define the inflammatory and angiogenic gene expression profile of whole blood cells at the site of occlusion in AMI patients.
Methods
The study was conducted in two stages (Figure 1). First, an array study was performed in five consecutive STEMI patients. Using custom-designed DNA array technology, differential expression of 221 different genes involved in angiogenesis, inflammation and atherosclerosis was probed in samples obtained at the site of coronary occlusion and peripheral circulation. Genes found to be differentially regulated were next validated in blood samples obtained in 28 distinct STEMI patients. Study protocols were approved by pertinent institutional ethics committees. Accordingly, informed consent was obtained from all patients.
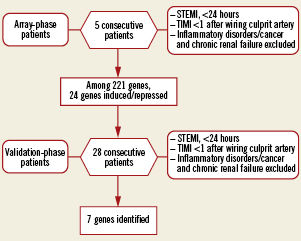
Figure 1. Flowchart of the study. Five consecutive patients were included in the array-phase study and 28 patients in the validation phase.
Patient selection for array study
We prospectively studied five consecutive patients with STEMI based on the following criteria: continuous chest pain suggestive of acute cardiac ischaemia, ST-segment elevation >1 mm in two contiguous leads for reperfusion with primary angioplasty and TIMI flow 0 (coronary occlusion) at coronary angiography. All subjects were older than 18 years of age. Patients with cardiac disease such as myocarditis, pericarditis and patients with chronic inflammatory diseases, malignant tumours, chronic renal failure on haemodialysis or infectious diseases were excluded.
Validation study participants
As a follow-up, 28 patients were enrolled in subsequent validation based on identical selection criteria as in the introductory array study. Six control patients were free of coronary artery disease.
Peripheral and local blood sampling
Blood samples were obtained during cardiac catheterisation as follows: The thrombotic occlusion in the culprit coronary artery was crossed with a 0.014 in. wire selected by the operator. Then, a 6Fr Export® AP Aspiration Catheter (crossing profile, 0.068 in.; Medtronic, Inc., Santa Clara, CA, USA) was advanced into the target coronary segment. Aspiration was performed under continuous negative pressure starting at the level of the thrombotic occlusion and advancing downstream into the target vessel. Blood was then filtered in order to eliminate thrombus and atherosclerotic debris. Peripheral blood was obtained through the guiding catheter positioned within the aorta. Blood for RNA analysis was collected into the Paxgene tube (BD Diagnostics, Franklin Lakes, NJ, USA). PCI was performed according to evidence-based guidelines. Gene expression analysis evaluated transcriptional expression dynamics between local and systemic samples.
RNA extraction
Total RNA was isolated from 1 ml of whole blood using a QIAamp RNA Blood Mini Kit (QIAGEN GmbH, Hilden, Germany). Genomic DNA contamination was eliminated by DNase treatment using TURBO DNA-free™ kit (Ambion/Applied Biosystems, Austin, TX, USA).
Array hybridisation
Gene expression profile was analysed using RT2 Profiler™ PCR Arrays (SABiosciences, Frederick, MD, USA). Data analysis was performed using an integrated web-based software package for the PCR Array system using ΔΔCt based fold-change calculations.
Selection of genes for validation
Based on results of the RT2 Profiler PCR array, a number of genes were chosen for validation. Selection criteria were based on the ratio of gene expression in local versus peripheral samples set at more than 1.5-fold increased/decreased in at least three out of five patients evaluated in the introductory array study.
Real-time polymerase chain reaction
Total RNA, extracted from whole blood, was reverse transcribed with random primers using the High-Capacity cDNA Archive System (Applied Biosystems, Foster City, CA, USA). Real-time RT-PCR on cDNA was performed using Assay-on-Demand (CCL2, FN1, CDH5, CCL18, SPP1 and CXCL12) or specific primers and TaqMan MGB probes (APOE-F: 5’-AGGAGCCGACTGGCCAAT-3’, APOE-R 5’-TCTGTCTCCACCGCTTGCT-3’, APOE-probe 6-FAM-TGGTCACATTCCTGGCAGGATGCC-MGB) in 96-well plates (ABI Prism 7000 Sequence Detection System; Applied Biosystems, Foster City, CA, USA) following the scheduled program (2 min 50°C, 10 min 95°C and 15 sec 95°C, 1 min 60°C for 40 cycles). Analysis was performed in triplicate. The relative expression of CCL2, FN1, CDH5, CCL18, SPP1, CXCL12 and APOE was normalised to the amount of GAPDH (Applied Biosystems, Foster City, CA, USA) in the same sample.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.00 (GraphPad Software, Inc., San Diego, CA, USA). Continuous variables were presented as mean±SEM. Categorical variables are reported as frequencies and percentages. Comparison between local blood sampling and peripheral blood sampling was performed using a paired T-test for STEMI patients and with an unpaired T-test when comparing STEMI patients with controls. Comparison between continuous variables was performed using unpaired T-test. p<0.05 was considered statistically significant. In the present study, the sample size was estimated based on the smallest statistical difference found in the array-phase study (CDH5) where at least 28 patients were required to be included in order to detect >5X difference in the mRNA level as measured by qPCR (alpha 0.05, statistical power 0.8)
Results
Patient characteristics
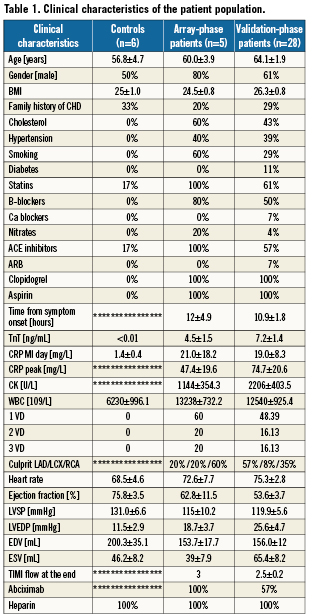
Demographics and clinical characteristics are shown in Table1. The array study was conducted in five patients (four males and one female). Two patients presented with late myocardial infarction (between 12-24 hours) and three with early myocardial infarction (<12 hours). The validation study was conducted with 28 patients. Among these patients, 51% were early myocardial infarction presentations (<12 hours).
Array study
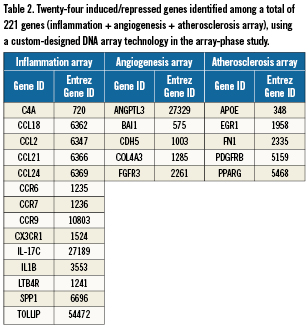
To study the gene expression in whole blood, the local sample (aspiration through the aspiration catheter at the site of occlusion) and the peripheral sample (aspiration through the guiding catheter in the aorta) were analysed using three different arrays totalling 221 genes belonging to families implicated in atherothrombosis. The first array focused on genes involved in angiogenesis (84 genes available at http://www.sabiosciences.com/rt_pcr_product/HTML/PAHS-024A.html), the second array focused on genes involved in inflammation (84 genes available at http://www.sabiosciences.com/rt_pcr_product/HTML/PAHS-011A.html), and the last array focused on genes involved in atherosclerosis (84 genes available at http://www.sabiosciences.com/rt_pcr_product/HTML/PAHS-038A.html). Among these genes, 24 were identified as differentially expressed with 1.5-fold induction or repression when comparing mRNA levels in the local with the peripheral blood sample (Table 2).
Validation study
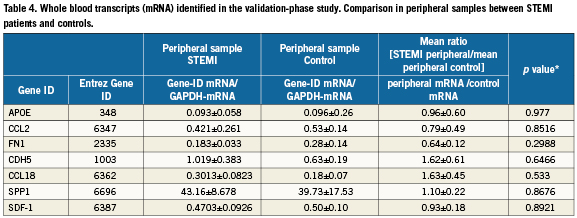
In the validation study, the mRNA levels of the 24 differentially expressed genes identified in the array study were studied using quantitative RT-PCR in 28 consecutive patients. Among them, seven genes showed significant differences between local mRNA expression and peripheral mRNA expression (Table3). The ratio (local mRNA/peripheral mRNA) were 223.4±82.9 for APOE (p=0.0172), 22.5±11.5 for CCL2 (p=0.0131), 36.4±7.6 for FN1 (p=0.0188), 7.0±1.6 for CDH5 (p=0.0138), 57.7±17.4 for CCL18 (p=0.0137), 12.21±5.7 for SPP1 (p=0.0076) and 15.0±3.8 for CXCL12 (p=0.03). Note, among these seven genes, no difference was noted in peripheral whole blood gene expression between the STEMI patients and controls (Table 4).
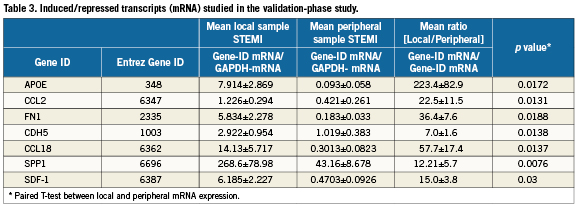
Network-based analysis of induced genes
In silico analysis of the seven significantly induced genes generated a non-stochastic network demonstrating activation of a G-protein/MAP kinase signalling program, and was determined to be non-arbitrary with a p-value=1.6×10–6 and a Z-Score of 60 (Figure 2A). Furthermore, GeneGo-based analysis ranked infarction, myocardial infarction and atherosclerosis as the top three pathological processes (Figure 2B), with NOTCH-induced endothelial-mesenchymal transition (EMT; critical for cardiac cushion development and response to myocardial injury) identified as the most highly induced pathway (Figure 2C).
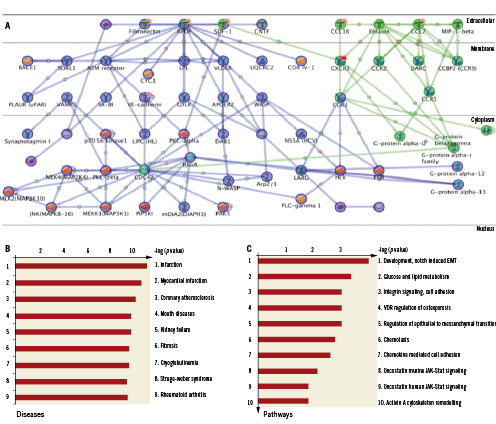
Figure 2. Induced coronary genes highly correlate with response to myocardial injury. A: Non-stochastic network generated from the seven induced genes (highlighted with orange circle on the network) identifies activation G-Protein and MAPK mediated signalling. B: Analysis of the Gene-GO database on pathological processes ranked infarction, myocardial infarction and atherosclerosis as the top three processes, with NOTCH-induced endothelial-mesenchymal transition (EMT) highlighted as the most highly induced pathway, C.
Local/peripheral RNA ratio of induced genes and myocardial infarction size
Among seven induced genes, only CCL2, highly correlated with G-protein, mediated signalling (Figure 2A), and showed a direct relationship with infarct size. Peak creatine kinase was significantly lower in the 1st tertile of CCL2 mRNA ratio (local/peripheral) as compared to 3rd CCL2 tertile (1472±304.9 vs.3468±841.4, p<0.05) (Figure3). A similar trend was noted for peak troponin T level (1st tertile vs. 3rd tertile [5.110±1.598 vs.12.27±3.036], p<0.053). No relationship with other clinical, haemodynamic or humoral parameters for CCL2 or other genes was noted.
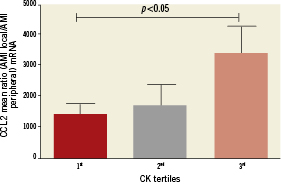
Figure 3. Peak creatine kinase was significantly lower in the 1st tertile of CCL2 mRNA ratio (local/peripheral) as compared to the 3rd CCL2 tertile (1472±304.9 vs. 3468±841.4, p<0.05)
Discussion
This study investigates the whole blood gene expression of genes belonging to the family of critical signalling pathways involved in the pathogenesis of plaque rupture and thrombosis in acute myocardial infarction. The PCR array technology was used to analyse 221 genes involved in inflammation, angiogenesis and atherosclerosis. Our results highlight the induction of transcripts belonging to the chemokine family, in particular CCL2 (MCP-1), CCL18 and CXCL12 (SDF-1), genes involved in the cell-cell or cell-extracellular matrix interaction family, namely FN1 (fibronectin 1), CDH5 (VE-cadherin) and SPP1 (osteopontin), as well as APOE (apolipoprotein E), at the site of occlusion during myocardial infarction. Furthermore, the ratio local/peripheral CCL2 gene expression was significantly associated with a higher myocardial infarction extent.
CCL2, CCL18 and CXCL12
Chemokines are chemotactic cytokines critical for leukocytes homing9. Although numerous experimental studies have shown an important role of chemokines in the pathogenesis of atherosclerosis, their role in the acute phase of myocardial infarction in humans is not well understood10. Here, we show that among 42 tested chemokines and chemokines receptors (there are ~50 cloned chemokines and ~20 chemokines receptors), CCL2 (MCP-1), CXCL12 (SDF-1) and CCL18, in particular, were highly induced in whole blood at the site of acute coronary occlusion.
CCL2 (MCP-1) belongs to the most studied chemokine member in cardiovascular disease. It has a pleiotropic role in atherosclerosis11 being central in lesion leukocyte infiltration and neointimal smooth muscle cell expansion12. Our finding of induced CCL2 expression at the site of the occlusion being proportional to the size of infarction further underlines its potential role also in the pathophysiology of acute coronary occlusion during myocardial infarction. The present finding also provides a mechanistic support to the data obtained from the A to Z trial, where plasma level of CCL2 showed an independent prognostic value in the acute and chronic phase of the acute coronary syndrome13,14. While CCL2 is central to alterations in plaque composition, CXCL12 (SDF-1) is critically involved in the endogenous repair and healing processes after myocardial damage15. Though the CCL2 expression is well documented at the myocardial level, controversial data were reported on the systemic levels of CXCL12 (SDF-1) in the acute phase of myocardial infarction16,17. The discrepancies were attributed to differences in the sampling sites, timing of sampling or differences in the therapeutic regimen. Our approach of analysing samples at the local and systemic sites at the time of reperfusion avoids these pitfalls. In fact, paired analysis showed a 15-fold increase of mRNA CXCL12 (SDF-1) levels between the site of occlusion as compared to peripheral samples, and for the first time demonstrates induction of the reparative homing signalling at the site of the coronary occlusion.
Similar to CCL12, whole blood cells depicted an upregulated chempokine CCL18 at the site of coronary occlusion. This chemokine is involved in the physiological homing of lymphocytes and dentritic cells as part of the primary immune response18, and was also found to be elevated in the plasma of patients with unstable angina19. Taken together, our data on the local upregulation of CCL2, CCL12 and 18 transcripts are consistent with the role of white blood cells in initiating the immediate immune response and reparative cascade at the time of thrombotic coronary plaque rupture.
FN1, CDH5 and SPP1
The extracellular matrix (ECM) is critical to maintain the structural integrity of the heart after myocardial infarction. Fibronectin and transforming growth factor (TGF)-β participate in transforming the fibroblasts into myofibroblasts, the main collagen-secreting cells at the healing site20. Other ECM proteins, such as thrombospondin-1 and -2, osteopontin, tenascin-C, periostin, secreted protein acidic and rich in cysteine (SPARC), promote matrix organisation in the infarcted myocardium21.
Among all these players, FN1 (fibronectin) and SPP1 (osteopontin) were locally induced at the site of coronary occlusion in the current study. The finding of a rapid induction of FN1 expression at the vascular site of coronary occlusion is novel and extends the previous data reporting positive fibronectin staining during the first day after myocardial infarction monitored by immunohistochemistry22. Likewise, upregulation of whole blood SPP1 (osteopontin) in our study at the site of the plaque rupture was not known, and indicates that at least part of the SPP1 protein level found in the plasma of patients with acute coronary syndromes22 may originate from circulating blood cells. Finally, CDH5 or VE-cadherin, belongs to the cadherin family of cell adhesion molecules and its plasma levels are increased during myocardial infarction23. Our observation of elevated gene expression at the site of coronary occlusion is potentially important for a number of reasons. It supports its role in vascular permeability and leukocyte extravasation24. This may be important not only for mediating plaque rupture, but also as shown by Wojakowski et al, CDH5 is, in particular, upregulated in a subset of peripheral blood cells, so –called very small embryonic-like stem cells25 underlying the importance of VE-cadherin in the endogenous post-injury response. In this regard, FX06 is a naturally occurring peptide, which is able to compete with E1 fragments of fibrin for binding to CDH5 and thereby acting as an anti-inflammatory molecule26. It is of note that the recent FIRE trial showed a beneficial effect of FX06 in the setting of myocardial infarction by reducing the necrotic core visualised by cardiac magnetic resonance27. Thus, the observation of elevated CDH5 levels at the site of coronary occlusion and the beneficial effects of FX06 on the necrotic core further support the concept of early leukocyte transendothelial migration in myocardial infarction mediating the early injury response28. In other words, circulating white blood cells accumulated at the site of the thrombotic plaque rupture transmigrate and contribute to the presence of extracellular matrix proteins such as fibronectin, osteopontin or VE cadherin in the early hours at the site of myocardial injury, and thereby initiate the process of matrix remodelling and infarction healing.
APOE
APOE (apolipoprotein E) is a well characterised lipoprotein involved in the clearance of triglyceride-rich lipoproteins in the liver. Early data suggested also a lipoprotein independent role of APOE in atherogenesis. In fact, APOE is expressed in various cells including hepatocytes, neuronal cells, smooth muscle cells or macrophages29. In macrophages, it exerts pleiotropic effects with anti-inflammatory30, antiproliferative31 and immunomodulatory downstream action32. The molecular mechanism of immunomodulatory effects in vivo remains unclear and is postulated to be exerted through toll-like receptors (TLR) 3 and 4 and expression of interleukin-1233. The marked upregulation of APOE expression at the site of the coronary occlusion was not previously described. Among all transcripts under study, APOE showed the highest local gene induction in whole blood cells. Given the multifaceted role of APOE in immunomodulation, this upregulation is striking and further experimental studies are needed to unravel the effects of this lipoprotein in the acute phase of myocardial infarction.
Limitations
This novel study focused on deciphering critical signalling pathways at the site of coronary plaque rupture in patients with acute myocardial infarction. As such, the study was primarily powered to identify differences in whole blood gene expression between local and peripheral samples obtained at the time of coronary reperfusion. This might have prevented recognition of differences in gene expression in peripheral samples between STEMI patients and controls in the later course post-infarction. Furthermore, only whole blood gene expression analysis was performed without discriminating different blood cell populations. Accordingly, our study does not provide the definitive cellular source of the gene expression pattern, which likely results from the predominant white blood cells. Nevertheless, the advantage is that it provides the signature of total blood mRNA underlying the protective or reparatory response regardless of the cell source in the setting of acute myocardial infarction. Finally, array analysis focused on the gene expression profile related to angiogenesis, inflammation and atherosclerosis as the main pathways involved in plaque rupture and initial healing response. Further studies using high throughput genomics or proteomics are needed to identify systemic molecular networks activated locally at the time of the occlusive coronary plaque rupture. It should also be noted that the study was not powered to address the prognostic importance of identified transcripts with regard to ventricular remodelling and function after successful reperfusion. In addition, the question whether differences in ischaemic time may affect gene expression of angiogenic or inflammatory genes needs to be addressed in the larger cohort of patients with varying ischaemic times at the time of the clinical presentation.
Conclusion and clinical implications
We identified a set of gene transcripts induced at the site of acute thrombotic coronary occlusion in whole blood during myocardial infarction. The results point at the predominant role of chemokines, cell-extracellular matrix and lipoprotein alterations in the pathophysiology of acute myocardial infarction and initial response to myocardial injury. The current study is also “hypothesis-generating” and identifies new white blood cells-related markers associated with acute coronary occlusion, which may serve as therapeutic targets in the novel pharmacological discovery using the local, stent-based drug delivery. Further experimental and clinical studies are needed in order to further decipher and validate the systemic molecular fingerprint at the site of coronary occlusion and its importance for the post injury remodelling after successful reperfusion, as well as prognostic value in post-infarction stratification.
Funding sources
Meijer Foundation for Cardiac Research, Aalst, Belgium.
Conflict of interest statement
The authors have no conflict of interest to declare.