
In the internet age, searching for information has become such a simple and speedy action as to deserve a neologism, “googling”. Every time we are frustrated by a general question (“what was the name of Napoleon’s horse?”) the easiest solution is to look for it on Google, which almost invariably refers us to the voice of the largest collaborative encyclopaedia in the world, Wikipedia. The action of consulting Google and Wikipedia through the screen of a smartphone has become so fast that it is difficult to remember how we managed before their existence. We looked at printed encyclopaedias, of course, or we asked someone more knowledgeable. Many are certainly still using those methods, but are they comparable to the comfort of having all the world’s information available at your fingertips?
The counterpart of Google, in the scientific world, is PubMed. According to Wikipedia, PubMed contains over 24 million bibliographic references derived from 5,200 biomedical journals. The articles available in the form of abstracts number about 17 million, while the review articles amount to 1.5 million and the articles available in free full text are about 3 million. Online since 1996, PubMed indexes the scientific literature available from 1949 up to today, and it does it for free – not a negligible detail. PubMed is in fact a search engine that draws on various databases of life sciences. These include the first database of the US National Library of Medicine, called MEDLINE.
Given the large amount of material indexed daily on PubMed, running a good search requires some priming information. Fellows are regularly amazed when they are introduced to the magic world of Boolean operators (“AND”, “OR” and “NOT” – three simple short words, and yet so powerful). I try to explain to them that a good search first and foremost needs the right dose of common sense. You simply cannot read everything; you need to identify the salient articles. Second, simply entering a jumble of keywords and clicking on one of the first results that is shown is typically a waste of time. Third, in addition to the Boolean operators, it is essential to use brackets, which makes the system understand the sequence by which the operators are applied. Once this basic information has been acquired, we are ready to progress to the next level, using filters and MeSH terms, not to mention the advanced search options. A smart fellow knows that a good literature search must not be limited to PubMed but should also be extended to careful scrutiny of the bibliography of each salient article.
Until recently I was convinced that the above methodology was the only solid one to provide a faithful snapshot of what is truly important on a given topic and, I must say, I felt sufficiently up to date compared with the old days when a trip to the university library was mandatory. A few weeks ago, however, I was chatting with a colleague who is an enthusiast about technology and I felt like Methuselah. We were discussing the left distal transradial approach (LDTRA), a vascular access technique about which a seminal article was recently published in EuroIntervention, authored by the universally acclaimed father of the radial approach, Ferdinand Kiemeneij1. Using his smartphone, my colleague proudly showed me the photo of his latest LDTRA case (a thumbs-up photo of a smiling patient with the sheath still inserted in his snuffbox), so I started challenging him with questions such as “In how many patients is it practicable?”, “In how many patients do you fail?”, “How do you close the vascular access at the end of the procedure?”. My colleague replied promptly, and revealed a fair number of tips and tricks that I had never read about. So I asked something like “How did you acquire all this knowledge in such a short time, considering that the LDTRA technique is in its infancy and there is still nothing (or next to nothing) in the literature?”. The answer was clear and destabilising: “I learned it on Twitter”.
Twitter-based learning, then. Amazed by this response, I decided to check for myself. A few days later, using the traditional approach, I performed a PubMed search using the following string: (ldTRA [Title/abstract] OR rdTRA [Title/abstract] OR snuffbox [Title/abstract] OR “distal radial” [Title]) AND coronary [Title/abstract]. The system provided me with 17 results, of which only 10 were more recent than the Kiemeneij paper. Of these 10 articles, one was not relevant, one was a technical report, five were small case series and three were single case reports. In this issue of EuroIntervention, we publish a new series of 200 patients treated with the LDTRA approach, the largest series reported so far2.
Then I repeated my search on Twitter, using the hashtags #ldtra (left distal transradial access) and #rdtra (right distal transradial access). The search using hashtags may not have the rigour and completeness of a PubMed search but it works pretty well, I must say. The first tweet I found chronologically was posted in June 2017 by an operator who attached four images of the LDTRA procedure. Following this first tweet, I then saw a group of similar cases performed by various interventionalists from all over the world, all accompanied by the inevitable “thumbs-up” photo. In their tweets, many addressed Dr Kiemeneij (@ferdikiem) directly, asking for advice. Someone thanked him for having “saved his back”. From July to October 2017, a lot of specific tweets began to appear, with short and snappy comments, questions, and answers. Why do you prefer the LDTRA? “More comfortable for the right-handed, more comfortable for the operator”, Kiemeneij tweeted in response to another user. Curiously, in July, the first photos of closure devices for the LDTRA also began to appear, first rudimentary and adapted, then dedicated and custom-made. At one point in time, the volume of tweets further increased, and the cases gradually became more complex – acute coronary syndromes, venous grafts, left main, left main treated by the double-kissing crush technique. Month after month, a chorus of enthusiasts emerged and the thumbs-up photos multiplied. In November 2017, the angioplasty.org account tweeted, “not sure, but I believe that #ldTRA may be the first major procedure in interventional cardiology that has been taught mainly via @Twitter”. The user @matheenkhuddus tweeted, “used 10 minutes manual compression with #StatSeal after #ldtra followed by 2 band-aids as done by @ferdikiem. Patient had a hard time understanding why he couldn’t leave immediately after”. In March 2018, I saw the first case of chronic total occlusion recanalisation with double distal radial access, right and left (two thumbs up). In May 2018 I even noted a flourish of proposals for improving the technique, e.g., “leave the sheath 10 cm out and taped on the hand. Very nice ergonomics, and easy to manage post as usual” (@RinfretStephane). The rest is the story of the times in which we live. I do not think I need to find a study in PubMed to convince me about the feasibility of the LDTRA in various scenarios now. It is all there, on Twitter. Of course, I will still need PubMed to convince me about success rates, efficacy, safety and that sort of thing. The article in this issue of EuroIntervention, reporting arterial puncture success in 96% of patients and puncture site complications in 8%, is a nice contribution to our understanding of this new approach2.
In another editorial, our Editor in Chief warns against the casual use of social media as a surrogate for traditional scientific debates3.
Indeed, an in-depth scientific reflection cannot easily be captured by a short, volatile tweet (Figure 1); however, Prof. Serruys, who has always been keen to promote innovation, is certainly aware of the huge potential of these new channels, especially when it comes to teaching tools and techniques. It is no coincidence that EuroIntervention has appointed its own Twitter editor, Salvatore Brugaletta4. In the case of the LDTRA, Twitter has caused this technique to go viral, has illustrated it with photos and real cases, has encouraged those who are fearful, and convinced some sceptics. How much more time would the traditional scientific literature take to bring this approach to the attention of the interventionists? Judging by the few articles on PubMed, the busy reader would be tempted to conclude that the LDTRA is currently used by only a few. Judging by Twitter, however, this technique already boasts a community of enthusiastic supporters, who exchange photos, experiences and information. Traditional publishing takes months to years for something to explode; Twitter takes minutes to days. Should we totally neglect the potential of #CardioTwitter in education?
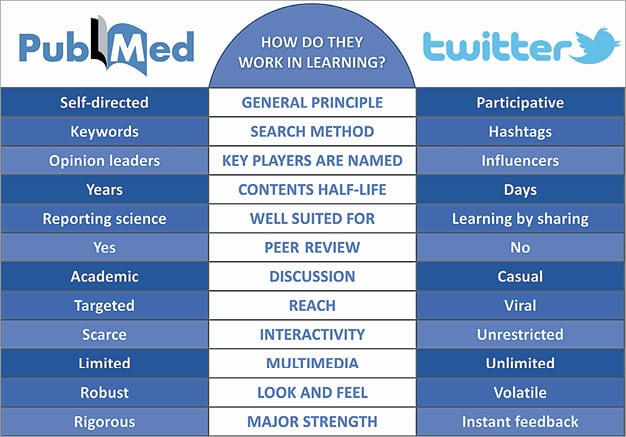
Figure 1. Characteristics of learning through the literature and the twitterature.
We previously discussed how the advent of new media represents a major opportunity in terms of multimedia and reach5. It is not difficult to envisage a future in which outcome data will be in scientific journals, while techniques, tutorials and clinical cases will be on the web, where they benefit the most from the unlimited multimedia opportunities represented by video and images, and the interactivity opportunity offered by social media. EuroIntervention and PCRonline (with their websites, but also their Twitter and Facebook accounts) should be seen as complementary in that regard, and more will come. Hashtags as keywords: there is a whole underground world of valuable contributions that many fail to recognise if they rely only on traditional channels. It is in this twitterature that pearls such as the LDTRA can be found.

